Results of the Survey on Needs for Data Standardisation and Databases in Molecular Biophysics
or
Are We “FAIR” in Molecular Biophysics
Motivation
There is a need for better scientific data management in molecular biophysics and a lack of data format standards and open access repositories for raw and processed data.
Therefore one of the MOSBRI tasks has an aim to improve biophysical data management. Several biophysical techniques will be selected to standardise data formats and tools, and to create a pilot open access database.
(FAIR principles = Findable + Accessible + Interoperable + Reusable)

The field of molecular biophysics encompasses several dozen techniques, with new methodologies appearing almost annually. Given this range of alternative methods for accomplishing similar research goals there is a high degree of variation in the level of usage of each method in the community. Most of the methods lack any accepted standards of data records and/or publicly accessible repositories or databases, and there are certainly no shared standards for reporting raw data or derived results. Researchers utilizing these techniques approach them for their research projects with varying expectations, levels of expertise, and have different personal needs for data archival. The principles of FAIR data management are also not generally known or accepted. The community of scientists utilizing these methods does not belong to one particular field of science or industry and includes specialists in biology, physics, chemistry, medicine, drug development and other fields.
WP4 aims to develop standardised format(s) for selected biophysical methods, tools for data curation, and a pilot database. In order to realize these tasks, mapping of the needs of the community in this area is a prerequisite. Mapping performed in the form of an electronic survey must take into account the variety of levels of knowledge, use, expectations, experience with databases and data management, and scientific backgrounds of the respondents.
The survey
An online survey was prepared in early 2022 and initially presented to the MOSBRI partners only, to allow refinement of the survey, before being opened publicly in February 2022. The survey consisted of 23 questions (see below) and primarily focused on the 30 biophysical techniques offered through MOSBRI. The survey was closed in mid-March 2022 having received 178 responses in total across the two rounds (MOSBRI and global).
The results show that there is a strong variation in the use and knowledge of the individual experimental techniques and of experience of data sharing, a generally low level of research data management at the respondents’ institutions, but also a recognized need for data sharing, format standardisation and repositories for many of the techniques listed. A significant number of respondents indicated their interest in collaborating in data standardisation. The results indicate the direction of future WP4 activities, highlighting needs for data standards and database development for specific biophysical methods used for protein characterisation and the study of biomolecular interactions.
Results of the survey
Presented below are a selection of the results obtained in the survey, while all results can be viewed in the documents linked at the bottom of the page.
- Questions in the survey
- How many people completed the survey and who were they?
- Experience with data deposition
- What techniques are used and how much?
- Where are the highest needs for data standardisation?
- Conclusions from the survey
- Final deliverable report and annex 1 documents and raw data
Questions in the survey
The survey questions were focused on identifying the expertise and scientific interests of the respondents, their use of techniques of molecular biophysics, views on the current situation and need for data format standardisation and needs for repositories or databases.
To view the full list of questions select “Questions asked in the survey” below:
Questions asked in the survey
About you
- What is your institution type?
- Your current position (or equivalent)
- Which field of research are you involved in?
- Are you involved in any particular area listed below?
- Are you a specialist in any of the following fields of structural biology?
- Have you ever deposited any scientific data into a public database/repository?
YES – Which database/repository?
NO – Is there a particular reason why not? - Do you have hands-on experience or have you used data from the following biophysical techniques?
- Do you have personal experience with any of the following aspects of molecular biophysics or structural biology data?
Measure/process/deposit - Do you regularly study/work with: Protein characterization/Nucleic acids, etc.
- Does your institution keep/support/participate in systematic long-term storage of scientific data?
(i.e. organized and searchable storage of raw and/or processed experimental data)
Could you provide us with further details?
Data standardisation
- Do you see the need for data standardisation in molecular biophysics?
- Do you see the need for applying the FAIR principles to biophysical data?
- Did you ever benefit from the FAIR type of access to scientific data (e.g. PDB, EMDB)?
Techniques used
- Identify techniques and frequency of usage in your projects (laboratory, group, close collaborator).
- In your experience/opinion, data from which techniques, measured in your projects, need to be accessed and should be therefore accessible for others?
Need for standardisation and databases
- In which area of data handling and processing have the following techniques the highest need for improvement
(what data handling aspects are not but should be covered)? - How high is the need for public and FAIR data repository/database for a given technique?
Extras
- If you were given access to original data from a biophysical technique, what information would make the data trustworthy for you?
(e.g. particular metadata, experimental condition, etc.)? - Do you know or can you estimate how many instrument manufacturers are there for a given technique worldwide?
- Do you have any experience with data management in biophysical techniques?
- Would you like to collaborate in developing data standards?
- Optional contact information
- Are you aware of any similar initiative regarding management of molecular biophysics data?
How many people completed the survey and who were they?
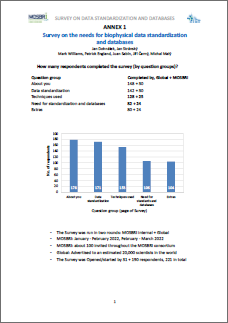
There were two rounds of the survey, first an internal round of the MOSBRI partners, then a global round. In total around 20 000 potential respondents were contacted.
| Question Group | Completed by Global + MOSBRI |
||
| About you | 148 + 30 | ||
| Data standardisation | 142 + 30 | ||
| Techniques used | 128 + 25 | ||
| Need for standardisation and databases | 82 + 24 | ||
| Extras | 80 + 24 |
In total 221 respondents started the survey, of which 178 completed the first group of questions (About you), and 104 completed the whole survey. Questions regarding the most important information from the point of view of this survey – Need for standardisation and databases – were answered by 106 respondents.
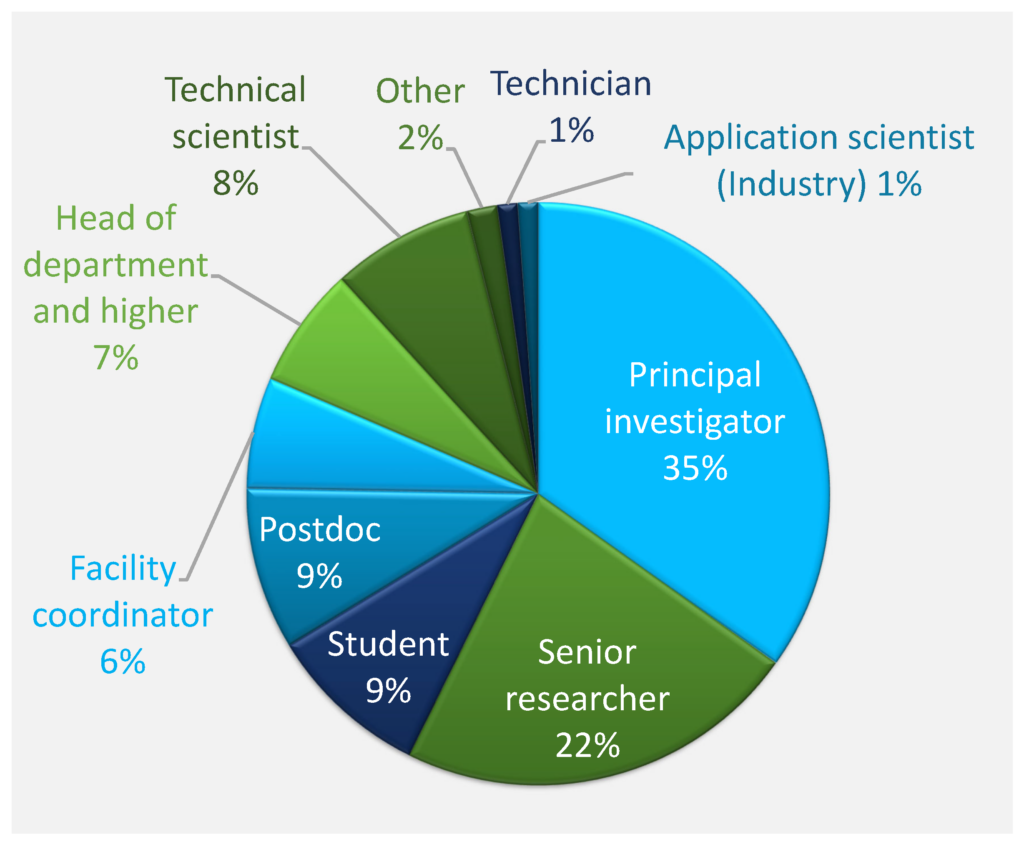
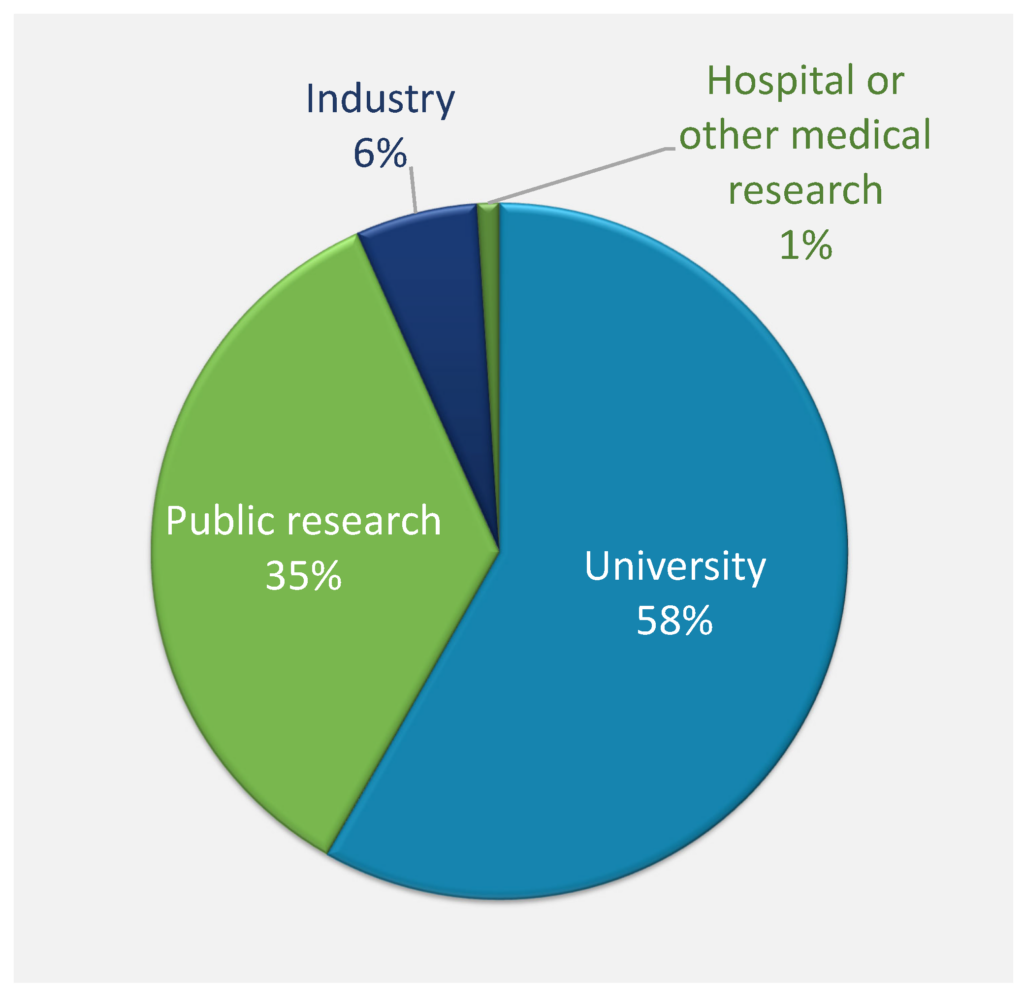
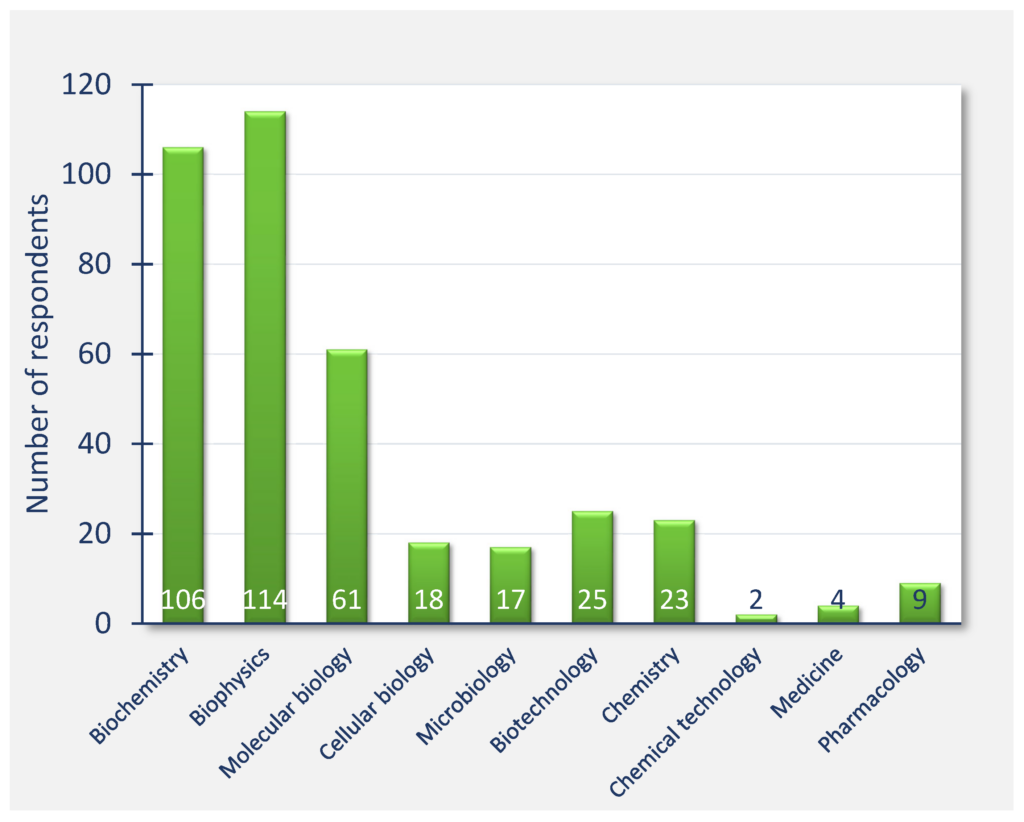
The survey respondents were mostly from universities and public research organizations (93%), while 6% were from industry. 64% of respondents belonged to the categories principal investigator, senior researcher, or head of department and higher, while the student and post‐doc categories represented 18% of respondents (Figure Qu. 2). Respondents were mostly active in the general fields of biophysics, biochemistry, and molecular biology, with the other fields represented by significantly smaller numbers (Figure Qu. 3). Responses to question 4 about narrower research areas showed that most of the respondents were mainly active in molecular biophysics, structural biology, and recombinant protein production.
The survey respondents mostly study proteins, protein‐protein interactions, and protein‐nucleic acid interactions, while the other types of biomolecules were significantly less represented.
Experience with data deposition
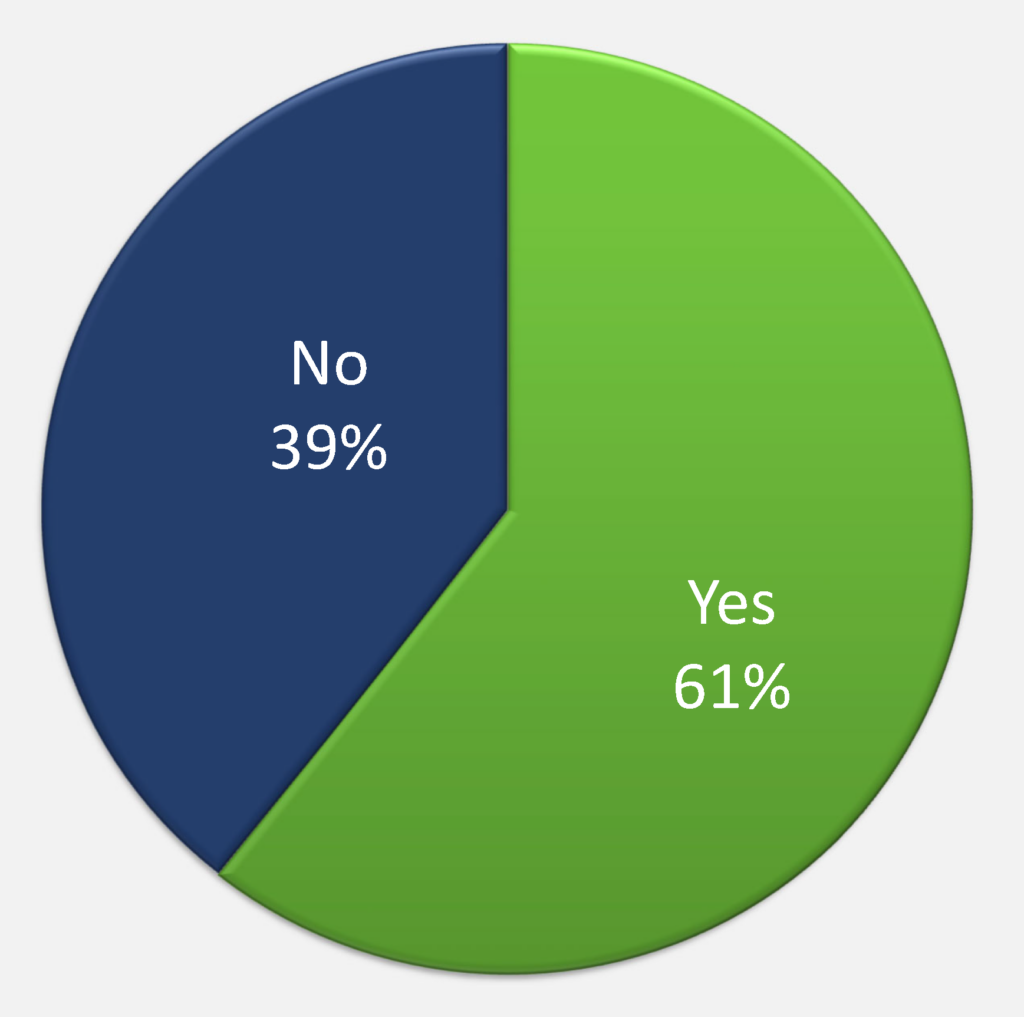
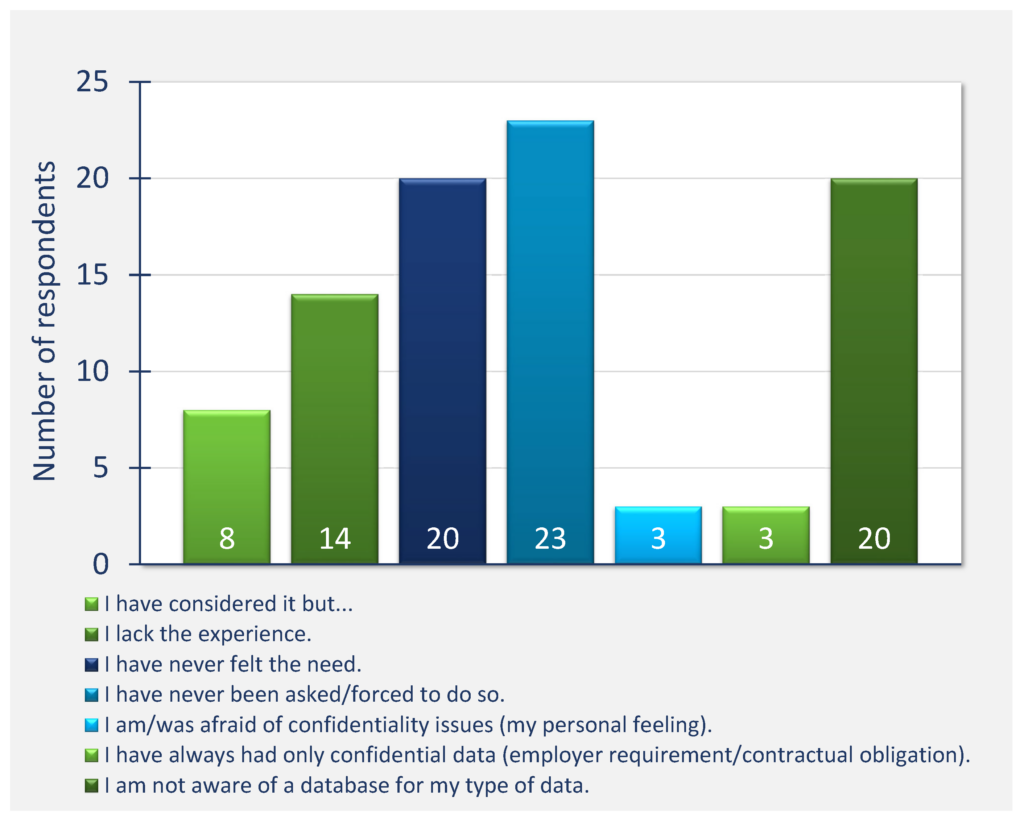
61% of respondents have had experience with data deposition into public databases or repositories, the PDB, SASBDB, and EMDB being mentioned most frequently. 49% of respondents have had experience with deposition of molecular biophysics and structural biology data (Question 8) and only 37% of respondents’ institutions supported systematic long-term storage of scientific data.
Figure Qu. 6: Have you ever deposited any scientific data into a public database/repository?
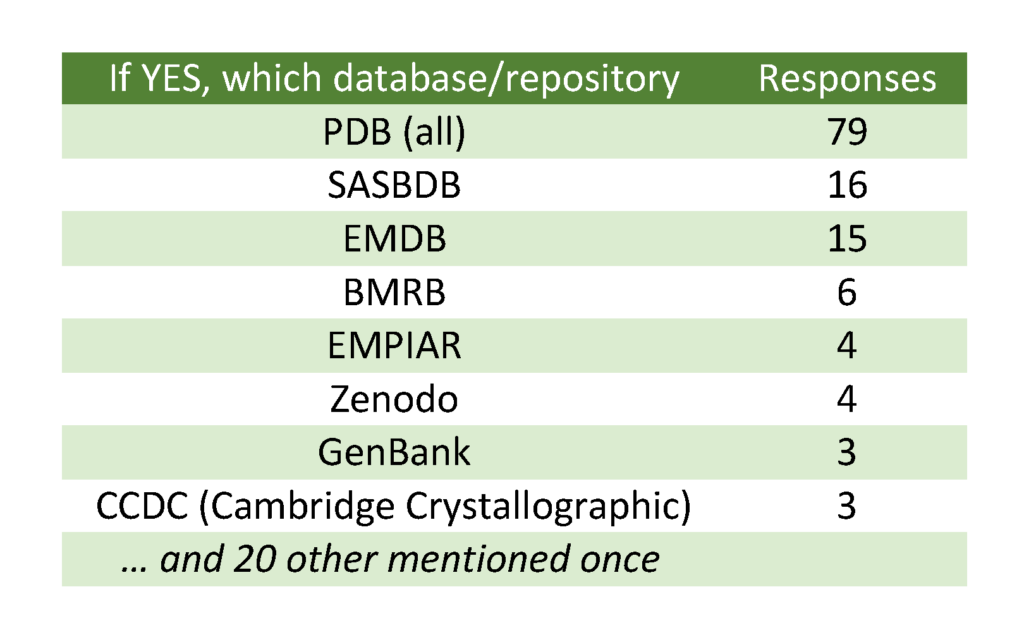
Several related efforts regarding management of biophysical data were mentioned by the respondents, mainly PCDDB, single crystal diffraction image repositories, CryoEM‐related efforts, MolMeDB, NMRlipids, an unspecified protein‐ligand binding database, and PDBDev (see Annex 1 for details).
What techniques are used and how much?
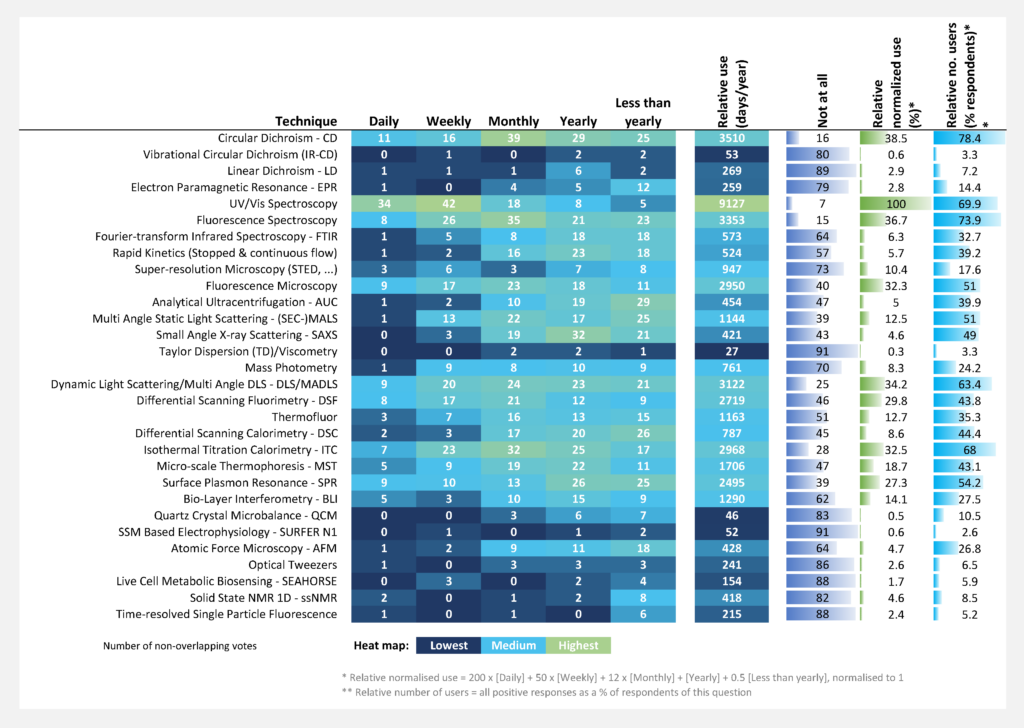
The techniques being used by the highest number of respondents are CD, UV/VIS spectroscopy, fluorescence spectroscopy, DLS, ITC, and SPR (Figure Qu. 14).
The techniques more heavily used (i.e., frequency of usage) are UV/VIS spectroscopy, CD, fluorescence spectroscopy, DLS, ITC, fluorescence microscopy, DSF, and SPR (Figure Qu. 14).
Where are the highest needs for data standardisation?
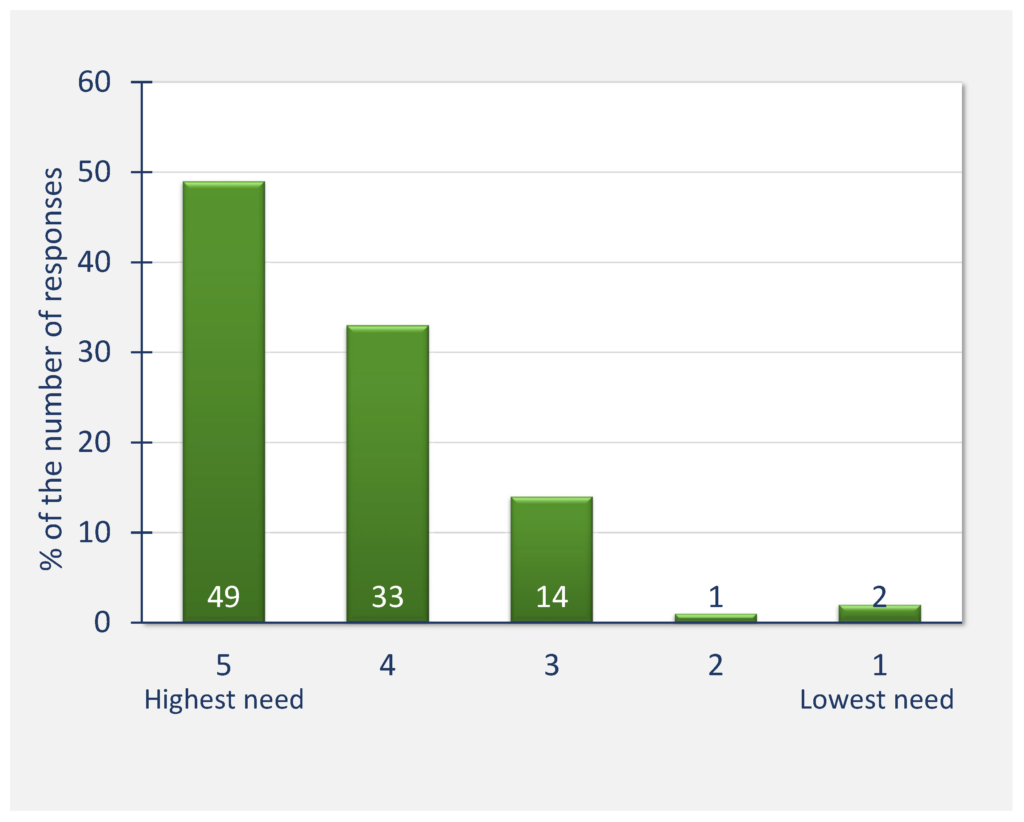
82% of all respondents see a high or relatively high need for data standardisation in molecular biophysics and a similar proportion perceive a high need for applying the FAIR principles in this field. Almost 90% of respondents acknowledge receiving benefit from access to scientific data made available according to FAIR principles.
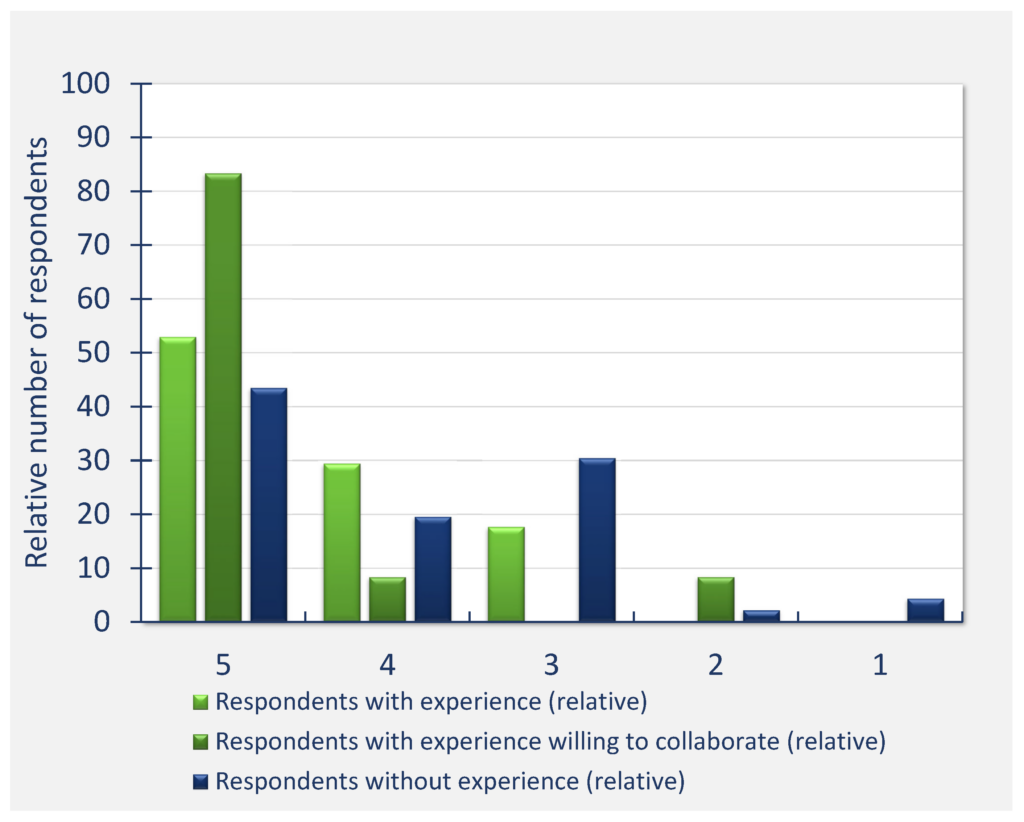
The highest perceived need for sharing data with the whole scientific community (Question 15, see Annex 1) was identified for SAXS (72% of respondents), ssNMR 1D (59.6%), super‐resolution microscopy (57.6%), time‐resolved single particle fluorescence (55.4%), and then other techniques.
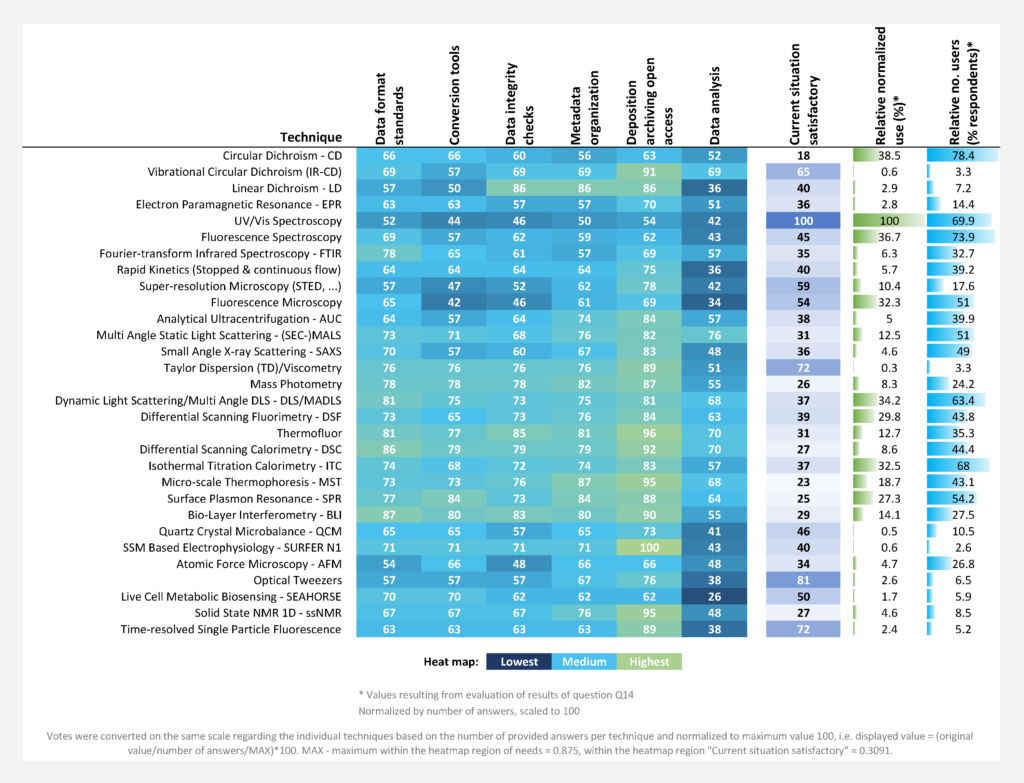
(Scaled to the number of answers per technique)
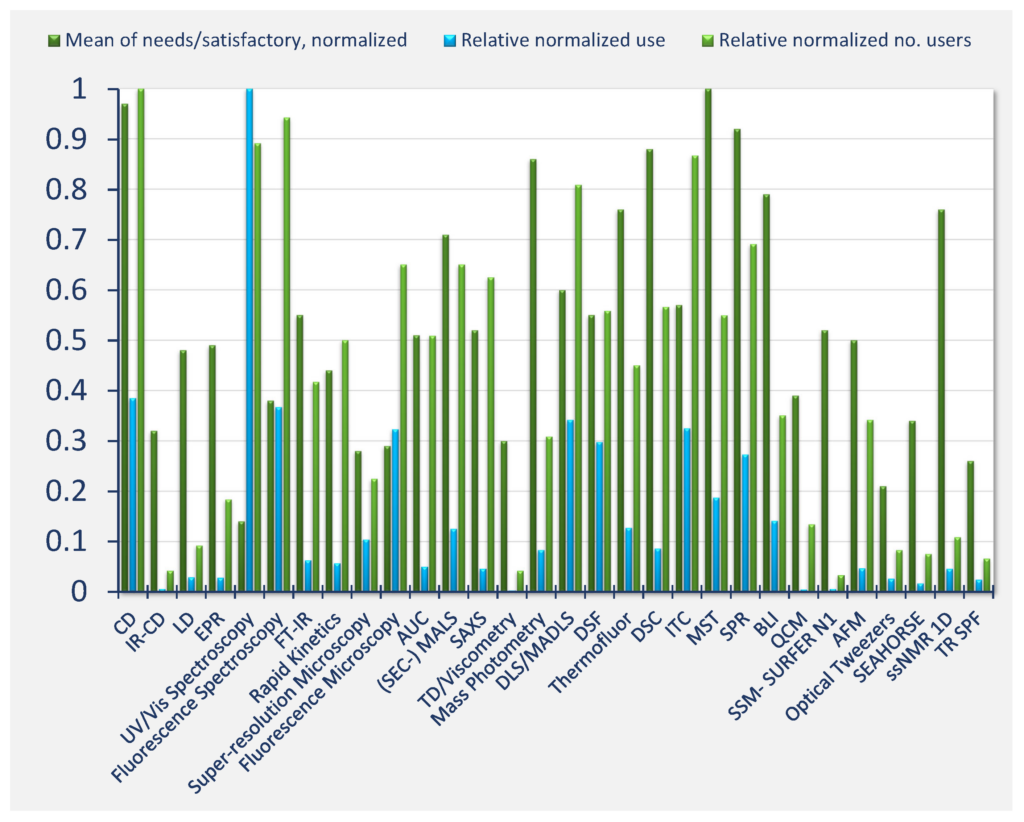
The highest need for development of data standards (mean over subcategories was identified for MST (relative normalized need 1.0), CD (0.97), SPR (0.92), DSC (0.88), mass photometry (0.86), BLI (0.79), thermofluor (0.76), ssNMR 1D (0.76), and (SEC)‐MALS (0.71) (Figure Qu. 16-2). The figure shows a combination of information from two separate questions. “Mean of needs/satisfactory, normalized” (dark green) is the average number of votes over the individual data management categories for a technique divided by the number of votes for “Current situation satisfactory” (Question 16), this distribution over all techniques was normalized to maximum value of 1. “Relative normalized use” (blue) is the Relative use in days per year, distribution normalized to maximum value 1 (see Question 14). “Relative normalized number of users” (light green) is the total number of votes indicating use of a technique, the distribution over all techniques was normalized to maximum value 1 (Question 14).
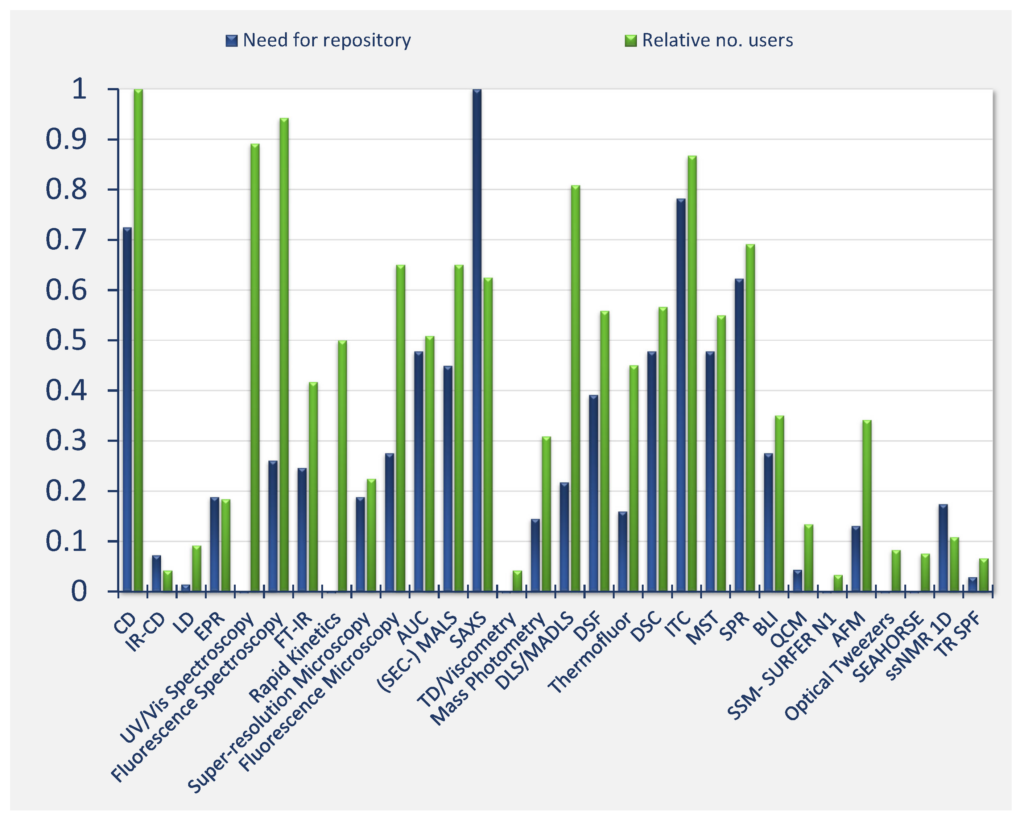
The respondents identified the highest needs of public and FAIR data repositories (weighted mean of answers to Question 17, Annex 1) for the following techniques: SAXS, ITC, AUC, DSC, SPR, CD, MST, (SEC)‐MALS, and ssNMR 1D.
Figure Qu. 17 shows normalised responses regarding the need for databases and normalized the number of users. “Need for repository” is the slope of linear regression of the number of votes indicated for need values 1‐low to 5‐high, normalized to maximum value 1.
The “Relative number of users” is calculated as in Figure Qu. 16. See the survey report (Annex 1) for further details.
Conclusions from the survey
The results of the survey on the needs for biophysical data standardisation and databases have shown the following points are important for the development of data standards, software tools, and a pilot database under WP4 of MOSBRI:
- The community of users of the techniques of molecular biophysics is well aware of the needs of FAIR data management in this field and indicated a high level of need for data format standardisation and repositories for a number of techniques.
- The relative use and number of users differ significantly across the individual techniques and this must be taken into account when focusing resources on particular techniques during the further activities of WP4.
- The following techniques (among those included in the survey) show frequent use or high numbers of users.
The top 10, shown in order from the highest, are: UV/VIS spectroscopy, CD, fluorescence spectroscopy, ITC, DLS/MADLS, fluorescence microscopy, SPR, DSF, MST, and DSC. - The following techniques (among those included in the survey) show a high need for data standards and for a repository
The top 10, shown in order from the highest mean of normalized needs for standards and databases, are: MST, CD, SPR, DSC, SAXS, ssNMR 1D, BLI, mass photometry, (SEC)‐MALS, and ITC. - The results of the survey have identified other important projects related to WP4 of MOSBRI and opened up a number of new contacts for potential collaboration.
- The survey results are of high value and will be fully exploited in further tasks under WP4.
Final deliverable report, annex 1 documents and raw data
The final report describing the work carried out and the summary of results of the survey, as well as the supporting Annex 1 document containing full analysis of the responses to the questions, can be downloaded below by clicking the filenames under the images.
Deliverable 4.2 Analysis of the mapping of the
needs for data standards/formats.

MOSBRI-Deliverable-D4_2-Survey_Biophysical-Data-Standards-and-Accessibility.pdf
Annex 1 Analysis of the mapping of the
needs for data standards/formats.
MOSBRI-Survey_Biophysical-Data-Standards-and-Accessibility_Annex1.pdf
The anonymized raw data from the survey have been deposited at Zenodo
(DOI 10.5281/zenodo.6604159).
List of abbreviations
| AFM | Atomic force microscopy |
| ARBRE-MOBIEU | Association of Resources for Biophysical Research in Europe – Molecular Biophysics in Europe |
| AUC | Analytical ultra-centrifugation |
| BLI | Bio-layer interferometry |
| CCPEM | Collaborative Computational Project for Electron cryo-Microscopy |
| CCP4 | Collaborative Computational Project no. 4 |
| CD | Circular dichroism |
| Cryo-EM | Electron cryo-microscopy |
| DLS | Dynamic light scattering |
| DMP | Data management plan |
| DSC | Differential scanning calorimetry |
| DSF | Differential scanning fluorimetry |
| EBSA | European Biophysical Societies’ Association |
| EMDB | Electron Microscopy Data Bank |
| EPR | Electron paramagnetic resonance |
| EOSC | European Open Science Cloud |
| FAIR | Findable, Accessible, Interoperable, Reusable |
| FTIR | Fourier-transform infrared spectroscopy |
| IR-CD | Vibrational circular dichroism |
| ITC | Isothermal titration calorimetry |
| IUPAB | International Union for Pure and Applied Biophysics |
| LD | Linear dichroism |
| MADLS | Multi-angle dynamic light scattering |
| MALS | Multi-angle light scattering |
| MolMeDB | Molecules on Membranes Database |
| MST | Microscale thermophoresis |
| NMRlipids | Nuclear Magnetic Resonance lipids databank |
| ssNMR 1D | Solid state nuclear magnetic resonance 1 dimensional |
| PCDDB | Protein Circular Dichroism Data Bank |
| PDB | Protein Data Bank |
| PDBDev | Prototype Archiving System for Integrative Structures |
| QCM | Quartz crystal microbalance |
| SAB | Scientific Advisory Board |
| SASBDB | Small Angle Scattering Biological Data Bank |
| SAXS | Small angle X-ray scattering |
| SEAHORSE | Live cell metabolic biosensing |
| SEC-MALS | Size exclusion chromatography-multi-angle light scattering |
| SPR | Surface plasmon resonance |
| SSM–SURFER N1 | SSM based electrophysiology – SURFER N1 |
| TD | Taylor dispersion |
| TNA | Transnational access |
| TR SPF | Time resolved single particle fluorescence |
| UV/VIS | Ultraviolet/visible light |